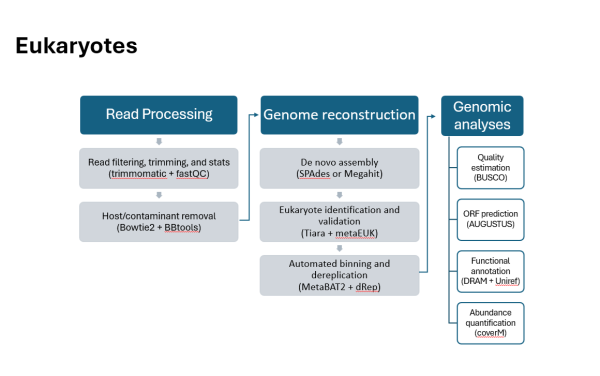
Eukaryotes Pipeline (Text)
- Read Processing
- Read filtering, trimming, and stats (trimmomatic + fastQC)
- Host/contaminant removal (Bowtie2 + BBtools)
- Genome reconstruction
- De novo assembly (SPAdes or Megahit)
- Eukaryote identification and validation (Tiara + metaEUK)
- Automated binning and dereplication (MetaBAT2 + dRep)
- Genomic analyses
- Quality estimation (BUSCO)
- ORF prediction (AUGUSTUS)
- Functional annotation (DRAM + Uniref)
- Abundance quantification (coverM)
