Microbiome Navigator
What is a microbiome navigator?
Our microbiome navigator is a microbiome science expert who can help:
- Provide guidance on microbiome science experimental design including sample collection and storage, identifying appropriate controls, selection of sequencing platform, and guidance on data processing and analyses
- Identify relevant resources for DNA/RNA extraction, library preparation, sequencing, and analysis of microbiome data
- Share information about working groups, courses, workshops, seminars, and socials that support trainees, faculty, and staff who are exploring microbiome science
How To Begin
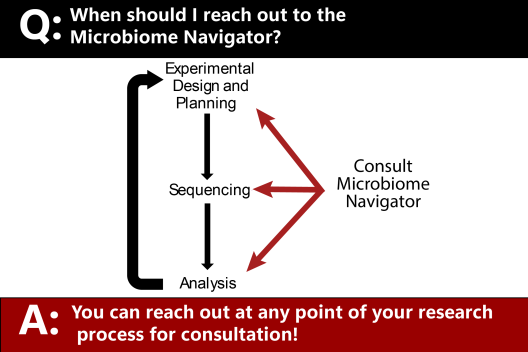
Contact our Microbiome Navigator by filling out the Intake Form.
First hour of consultation is free. Additional consulting is available at an hourly expense.
Get a quote to start work:
- Visit Questions and Quotes to start the quote process.
Who is our microbiome navigator?

Dr. Garrett Smith’s Background
I grew up in California, obtained my bachelors in Iowa, got my PhD at OSU in Microbiology in Kelly Wrighton’s laboratory, then did a postdoc in the Netherlands. My training exposed me to classical microbiology as well as cutting edge microbiome science in environmental and host-associated microbiomes. I have experience with microbial cultivation, chromatographs, 16S sequencing, metagenomes, metaproteomes, and metatranscriptomes. I got exposed to a lot of different kinds of frontier-expanding microbiome science throughout my training. For example, I’ve integrated taxonomic and functional information to generate novel ecosystem level insights on subjects ranging from seasonal effects on methane production in soil bacteria to amino acid fermentation in the mouse gut microbiome.
What makes you excited about each of your new roles as both Microbiome Navigator and Data Analyst within the Microbiome Platform at Ohio State University?
I really like bringing my microbiome science expertise to a broader audience both in my role as navigator and as a data analyst. I love hearing about the different types of science going on – from microbiology in food, to the space station, to agriculture, to health. I’ve always had an interest in where microbes are and what they’re doing. As a data analyst and the navigator, I can talk to researchers and help them identify the best approaches to answer their microbiome science science questions.
Besides microbes, what’s fun for you outside of work?
I like to be outside. I also read a lot of books and particularly enjoy science fiction. I’m currently reading the Imperial Radch series. I also like to hang out with family including my 2 year old and his 7 cousins who live in the area.
If an investigator is totally new to microbiome science, what kind of questions can you, as the Microbiome Navigator, help answer?
I can help you resolve which approaches, sequencing platforms, and analyses are most suitable for your sample type and project goals. I can walk through what a typical project might look like with various sequencing options, and I can help with your experimental design. I can help inform what is needed for an effective microbiome project, and identify additional resources on campus that may be useful to you.
What other resources on campus should investigators be aware of that may help them with projects that involve microbiome science?
- CoMS Working Groups – for help / guidance with microbiome analyses
- Microbiome Platform – For guidance on microbiome studies, DNA extraction / sequencing, or analysis support
- Center of Microbiome Science – additional microbiome science resources, faculty, courses, workshops, events etc.
Shared Resources
- CCTS (including Voucher Program for support toward Microbiome Platform sequencing or analysis)
- Campus Chemical Instrument Center - Metabolomics / Proteomics Core
In your role as a Data Analyst, you specialize in microbial metabolism. Why is microbial metabolism important for advancing microbiome science projects?
Microbes are more than just a taxon and fraction of your reads. They perform specific functions and sometimes a range of functions that can be turned on or turned off and can be critical for host response or an ecosystem process. We need to be able to identify these functions and associate them with the right taxa to help improve the conceptual models we develop to describe our ecosystem of interest.
What do you see as up and coming in microbiome science that you want our OSU community to be aware of?
The Data Analysts within the Microbiome Platform at Ohio State University are working on long read hybrid metagenomic assemblies and integrating data across sequencing platforms, so that we can get improved high quality genome recovery, including strain resolution. This will be a critical advance for situations like outbreak investigations, hard-to-culture infections, or the study of functions like antimicrobial resistance. We’re also working up metabolic modeling to better generate hypotheses to understand how microbes and microbial communities are interacting at gene and ecosystem levels. We’re applying advanced statistical analyses to connect microbiome structures to eco-systems and phenotypes. This helps us more effectively understand what microbes can do within a community.
Interview conducted by Vanessa Hale, Aug. 2024
The Microbiome Navigator is jointly supported by the Clinical and Translational Science Institute, Foods For Health, the College of Medicine, and the Center of Microbiome Science.
