Skip to main content
Ohio State navigation bar
Metabarcoding Pipeline
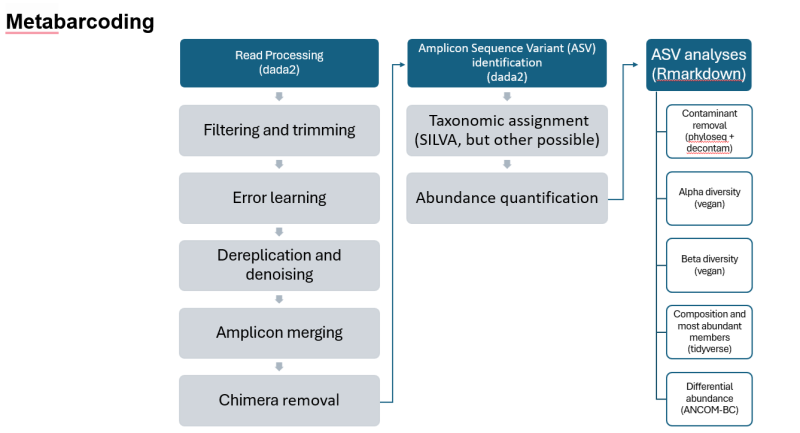
Metabarcoding Workflow
Metabarcoding Workflow (text version)
- Read Processing (DADA2)
- Filtering and trimming
- Error learning
- Dereplication and denoising
- Amplicon merging
- Chimera removal
- Amplicon Sequence Variant (ASV) identification (dada2)
- Taxonomic assignment (SILVA, but other possible)
- Abundance quantification
- ASV analyses (Rmarkdown)
- Contaminant removal (phyloseq + decontam)
- Alpha diversity (vegan)
- Beta diversity (vegan)
- Composition and most abundant members (tidyverse)
- Differential abundance (ANCOM-BC)