Skip to main content
Ohio State navigation bar
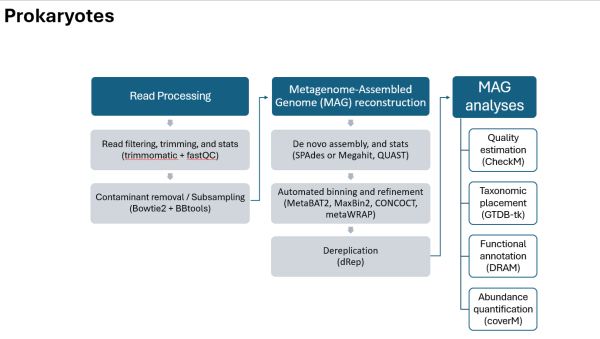
Prokaryotes Pipeline
Prokaryotes Workflow (text)
- Read Processing
- Read filtering, trimming, and stats (trimmomatic + fastQC)
- Contaminant removal / Subsampling (Bowtie2 + BBtools)
- Metagenome-Assembled Genome (MAG) reconstruction
- De novo assembly, and stats (SPAdes or Megahit, QUAST)
- Automated binning and refinement (MetaBAT2, MaxBin2, CONCOCT, metaWRAP)
- Dereplication (dRep)
- MAG analyses
- Quality estimation (CheckM)
- Taxonomic placement (GTDB-tk)
- Functional annotation (DRAM)
- Abundance quantification (coverM)